Spatial Predictions
Spatial Predictions
Introduction
The goal of this section is to demonstrate how to use models to predict back to spatial data or full image extents as opposed to individual image chips. To accomplish this task, I create a function that accepts a trained model object and an input file and returns a raster output with the same spatial extent and coordinate reference system as the original data and one channel storing the predicted class code. At the end of this module, I will also demonstrate writing all predicted class probabilities back to disk as a multiband raster grid.
Preparation
I begin by reading in the required libraries. I will use rasterio to read in the geospatial raster data. I will also make use of cv2 within the function for reading data. To display the resulting classification, I will use earthpy. I also set the device to the GPU to speed up inference.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import math
import os
import cv2
import torch
import torch.nn as nn
from torch.nn import functional as F
from torch.utils.data.dataset import Dataset
from torch.utils.data import DataLoader
import rasterio as rio
from rasterio.plot import show
import earthpy as epdevice = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
print(device)cuda:0I will use the model that was defined and trained in the last section using the Landcover.ai dataset. I first need to redefine the model architecture, instantiate an instance of the model, and move the model to the GPU. I then load the saved model parameters generated from the learning process from disk. These parameters have been provided with the class data if you did not run the training process demonstrated in the prior module.
def double_conv(inChannels, outChannels):
return nn.Sequential(
nn.Conv2d(inChannels, outChannels, kernel_size=(3,3), stride=1, padding=1),
nn.BatchNorm2d(outChannels),
nn.ReLU(inplace=True),
nn.Conv2d(outChannels, outChannels, kernel_size=(3,3), stride=1, padding=1),
nn.BatchNorm2d(outChannels),
nn.ReLU(inplace=True)
)def up_conv(inChannels, outChannels):
return nn.Sequential(
nn.ConvTranspose2d(inChannels, outChannels, kernel_size=(2,2), stride=2),
nn.BatchNorm2d(outChannels),
nn.ReLU(inplace=True)
)class myUNet(nn.Module):
def __init__(self, encoderChn, decoderChn, inChn, botChn, nCls):
super().__init__()
self.encoderChn = encoderChn
self.decoderChn = decoderChn
self.botChn = botChn
self.nCls = nCls
self.encoder1 = double_conv(inChn, encoderChn[0])
self.encoder2 = nn.Sequential(nn.MaxPool2d(kernel_size=2, stride=2),
double_conv(encoderChn[0], encoderChn[1]))
self.encoder3 = nn.Sequential(nn.MaxPool2d(kernel_size=2, stride=2),
double_conv(encoderChn[1], encoderChn[2]))
self.encoder4 = nn.Sequential(nn.MaxPool2d(kernel_size=2, stride=2),
double_conv(encoderChn[2], encoderChn[3]))
self.bottleneck = nn.Sequential(nn.MaxPool2d(kernel_size=2, stride=2),
double_conv(encoderChn[3], botChn))
self.decoder1up = up_conv(botChn, botChn)
self.decoder1 = double_conv(encoderChn[3]+botChn, decoderChn[0])
self.decoder2up = up_conv(decoderChn[0], decoderChn[0])
self.decoder2 = double_conv(encoderChn[2]+decoderChn[0], decoderChn[1])
self.decoder3up = up_conv(decoderChn[1], decoderChn[1])
self.decoder3 = double_conv(encoderChn[1]+decoderChn[1], decoderChn[2])
self.decoder4up = up_conv(decoderChn[2], decoderChn[2])
self.decoder4 = double_conv(encoderChn[0]+decoderChn[2], decoderChn[3])
self.classifier = nn.Conv2d(decoderChn[3], nCls, kernel_size=(1,1))
def forward(self, x):
#Encoder
encoder1 = self.encoder1(x)
encoder2 = self.encoder2(encoder1)
encoder3 = self.encoder3(encoder2)
encoder4 = self.encoder4(encoder3)
#Bottleneck
x = self.bottleneck(encoder4)
#Decoder
x = self.decoder1up(x)
x = torch.concat([x, encoder4], dim=1)
x = self.decoder1(x)
x = self.decoder2up(x)
x = torch.concat([x, encoder3], dim=1)
x = self.decoder2(x)
x = self.decoder3up(x)
x = torch.concat([x, encoder2], dim=1)
x = self.decoder3(x)
x = self.decoder4up(x)
x = torch.concat([x, encoder1], dim=1)
x = self.decoder4(x)
#Classifier head
x = self.classifier(x)
return xmodel = myUNet(encoderChn=[16,32,64,128], decoderChn=[128,64,32,16], inChn=3, botChn=512, nCls=5).to(device)saveFolder = "C:/myFiles/work/dl/landcoverai_models/"
best_weights = torch.load('data/models/landcoverai_unet_model.pt')
model.load_state_dict(best_weights)<All keys matched successfully>I will make a prediction to one of the provided images in the Landcover.ai dataset. I read in the image and display it using rasterio. I selected this image because it has a mix of different land cover types within the extent. Note that the markings along the margins are the coordinates relative to the coordinate reference system to which the data are referenced.
testImg = "data/files/N-33-60-D-c-4-2.tif"src = rio.open(testImg)
rio.plot.show(src)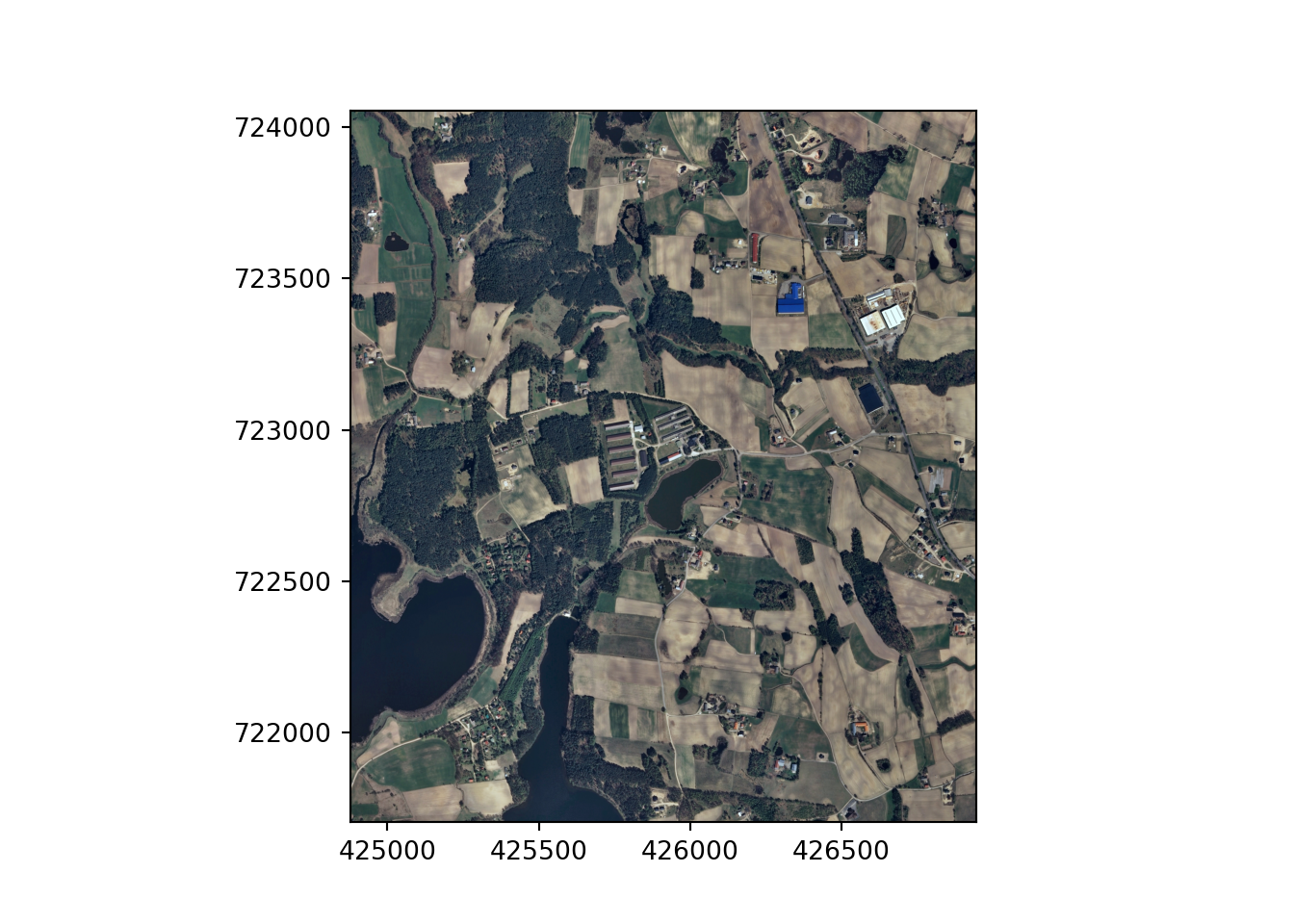
src.close()Make Classification Inference
I have defined a function below to make inferences to new data using a trained model. Here are descriptions of the input parameters.
- image_in: image to predict to. This image must have the same bands as the data used to train the model. The bands must also be in the same order as the original data. The data should be scaled using the same normalization parameters as were used in the training process.
- pred_out: name of file that will be written to disk plus the full path. Results are written as GeoTIF files with the same spatial reference information as the input image. The result will be a single band raster grid of class codes.
- model: model object that will be used to make predictions. You will need to instantiate an instance of a model and load the trained weights prior to making predictions.
- chip_size: size of image chips for processing. Although not strictly necessary, we recommend that this be the same as the chip size used to train the model.
- stride_x: stride in the x dimension (side-to-side).
- stride_y: stride in the y dimension (up-and-down).
- crop: number of columns or rows of pixels to crop from each side of the image chips while merging the output to a final product. This crop is not applied to margin chips so that the spatial extent of the output prediction is the same as the input image.
- n_channels: number of channels in the input data.
Before demonstrating this inference process, let’s discuss a few practical considerations. First, I have generally found that predictions near the edge of image chips are poorer than those in the interior. This may be due to not having a full set of neighbors when the convolution operations are applied. As a result, I recommend not using the edge pixels in the final merged output. This can be accomplished by (1) using a stride in the x and y directions that is smaller than the chip size so that each processed chip overlaps and (2) cropping the edge pixels. As noted above, as defined the function applies the defined cropping unless the chip is at the margin of the image. This is so that the spatial extent of the input image is maintained.
You may also find that very large images need to be broken into multiple smaller images, processed through the inference process separately, then merged to a final product. Images can be broken into smaller extents or tiles and the results can be merged to a final raster grid using GIS software tools, such as ArcGIS Pro or QGIS.
def geoInfer(image_in, pred_out, chip_size, stride_x, stride_y, crop, n_channels):
#Read in topo map and convert to tensor===========================
image1 = cv2.imread(image_in)
image1 = cv2.cvtColor(image1, cv2.COLOR_BGR2RGB)
image1 = image1.astype('uint8')
image1 = torch.from_numpy(image1)
image1 = image1.permute(2, 0, 1)
image1 = image1.float()/255
t_arr = image1
#Make blank grid to write predictions two with same height and width as topo===========================
image2 = cv2.imread(image_in)
image2 = cv2.cvtColor(image2, cv2.COLOR_BGR2RGB)
image2 = image2.astype('uint8')
image2 = torch.from_numpy(image2)
image2 = image2.permute(2, 0, 1)
image2 = image2.float()/255
p_arr = image2[0, :, :]
p_arr[:,:] = 0
#Predict to entire topo using overlapping chips, merge back to original extent=============
size = chip_size
stride_x = stride_x
stride_y = stride_y
crop = crop
n_channels = n_channels
across_cnt = t_arr.shape[2]
down_cnt = t_arr.shape[1]
tile_size_across = size
tile_size_down = size
overlap_across = stride_x
overlap_down = stride_y
across = math.ceil(across_cnt/overlap_across)
down = math.ceil(down_cnt/overlap_down)
across_seq = list(range(0, across, 1))
down_seq = list(range(0, down, 1))
across_seq2 = [(x*overlap_across) for x in across_seq]
down_seq2 = [(x*overlap_down) for x in down_seq]
#Loop through row/column combinations to make predictions for entire image
for c in across_seq2:
for r in down_seq2:
c1 = c
r1 = r
c2 = c + size
r2 = r + size
#Default
if c2 <= across_cnt and r2 <= down_cnt:
r1b = r1
r2b = r2
c1b = c1
c2b = c2
#Last column
elif c2 > across_cnt and r2 <= down_cnt:
r1b = r1
r2b = r2
c1b = across_cnt - size
c2b = across_cnt + 1
#Last row
elif c2 <= across_cnt and r2 > down_cnt:
r1b = down_cnt - size
r2b = down_cnt + 1
c1b = c1
c2b = c2
#Last row, last column
else:
c1b = across_cnt - size
c2b = across_cnt + 1
r1b = down_cnt - size
r2b = down_cnt + 1
ten1 = t_arr[0:n_channels, r1b:r2b, c1b:c2b]
ten1 = ten1.to(device).unsqueeze(0)
model.eval()
with torch.no_grad():
ten2 = model(ten1)
m = nn.Softmax(dim=1)
pr_probs = m(ten2)
ten_p = torch.argmax(pr_probs, dim=1).squeeze(1)
ten_p = ten_p.squeeze()
#print("executed for " + str(r1) + ", " + str(c1))
if(r1b == 0 and c1b == 0): #Write first row, first column
p_arr[r1b:r2b-crop, c1b:c2b-crop] = ten_p[0:size-crop, 0:size-crop]
elif(r1b == 0 and c2b == across_cnt+1): #Write first row, last column
p_arr[r1b:r2b-crop, c1b+crop:c2b] = ten_p[0:size-crop, 0+crop:size]
elif(r2b == down_cnt+1 and c1b == 0): #Write last row, first column
p_arr[r1b+crop:r2b, c1b:c2b-crop] = ten_p[crop:size+1, 0:size-crop]
elif(r2b == down_cnt+1 and c2b == across_cnt+1): #Write last row, last column
p_arr[r1b+crop:r2b, c1b+crop:c2b] = ten_p[crop:size, 0+crop:size+1]
elif((r1b == 0 and c1b != 0) or (r1b == 0 and c2b != across_cnt+1)): #Write first row
p_arr[r1b:r2b-crop, c1b+crop:c2b-crop] = ten_p[0:size-crop, 0+crop:size-crop]
elif((r2b == down_cnt+1 and c1b != 0) or (r2b == down_cnt+1 and c2b != across_cnt+1)): # Write last row
p_arr[r1b+crop:r2b, c1b+crop:c2b-crop] = ten_p[crop:size, 0+crop:size-crop]
elif((c1b == 0 and r1b !=0) or (c1b ==0 and r2b != down_cnt+1)): #Write first column
p_arr[r1b+crop:r2b-crop, c1b:c2b-crop] = ten_p[crop:size-crop, 0:size-crop]
elif (c2b == across_cnt+1 and r1b != 0) or (c2b == across_cnt+1 and r2b != down_cnt+1): # write last column
p_arr[r1b+crop:r2b-crop, c1b+crop:c2b] = ten_p[crop:size-crop, 0+crop:size]
else: #Write middle chips
p_arr[r1b+crop:r2b-crop, c1b+crop:c2b-crop] = ten_p[crop:size-crop, crop:size-crop]
#Read in a GeoTIFF to get CRS info=======================================
image3 = rio.open(image_in)
profile1 = image3.profile.copy()
image3.close()
profile1["driver"] = "GTiff"
profile1["dtype"] = "uint8"
profile1["count"] = 1
profile1["PHOTOMETRIC"] = "MINISBLACK"
profile1["COMPRESS"] = "NONE"
pr_out = p_arr.cpu().numpy().round().astype('uint8')
#Write out result========================================================
with rio.open(pred_out, "w", **profile1) as f:
f.write(pr_out,1)
torch.cuda.empty_cache()I am now ready to use the function to performance inference on the image. I am using a chip size of 512-by-512 pixels since this was the size used to train the model. I am using a stride of 256 pixels in the x and y directions to allow for a 50% overlap between adjacent processing chips. I am setting the crop parameters to 50 so that the outer 50 pixels of each chip are not included in the final output unless the chip occurs on the edge of the image.
geoInfer(image_in=testImg,
pred_out="data/files/example_pred.tif",
chip_size=512, stride_x=256, stride_y=256, crop=50, n_channels=3)Once the inference process executes, I read the result back in from disk and display the categorical raster using matplotlib and earthpy. The link provided as a comment provides examples of how to use earthpy to visualize raster data. Generally, the result looks pretty good. You may want to open the file in a GIS software to explore it further and compare it to the associated image.
#https://www.earthdatascience.org/courses/use-data-open-source-python/intro-raster-data-python/raster-data-processing/classify-plot-raster-data-in-python/
import earthpy.plot as ep
from matplotlib.colors import ListedColormap, BoundaryNorm
clsOut = rio.open("data/files/example_pred.tif")
clsOutA = clsOut.read().squeeze()
src.close()
print(np.max(clsOutA.shape))4701classes = list(np.unique(clsOutA).astype('int'))
classColors = ['gray', 'red', 'green', 'blue', 'black']
classNames = ['background', 'building', 'woodlands', 'water', 'road']
clsBounds = BoundaryNorm([-.5, .5, 1.5, 2.5, 3.5, 4.5], len(classes))
colorMap = ListedColormap(classColors)
f, ax = plt.subplots(figsize=(10,5))
im = ax.imshow(clsOutA, cmap = colorMap, norm=clsBounds)
ax.set(title="Landcover classes")
ep.draw_legend(im, titles = classNames, classes = classes)
ax.set_axis_off()
plt.show()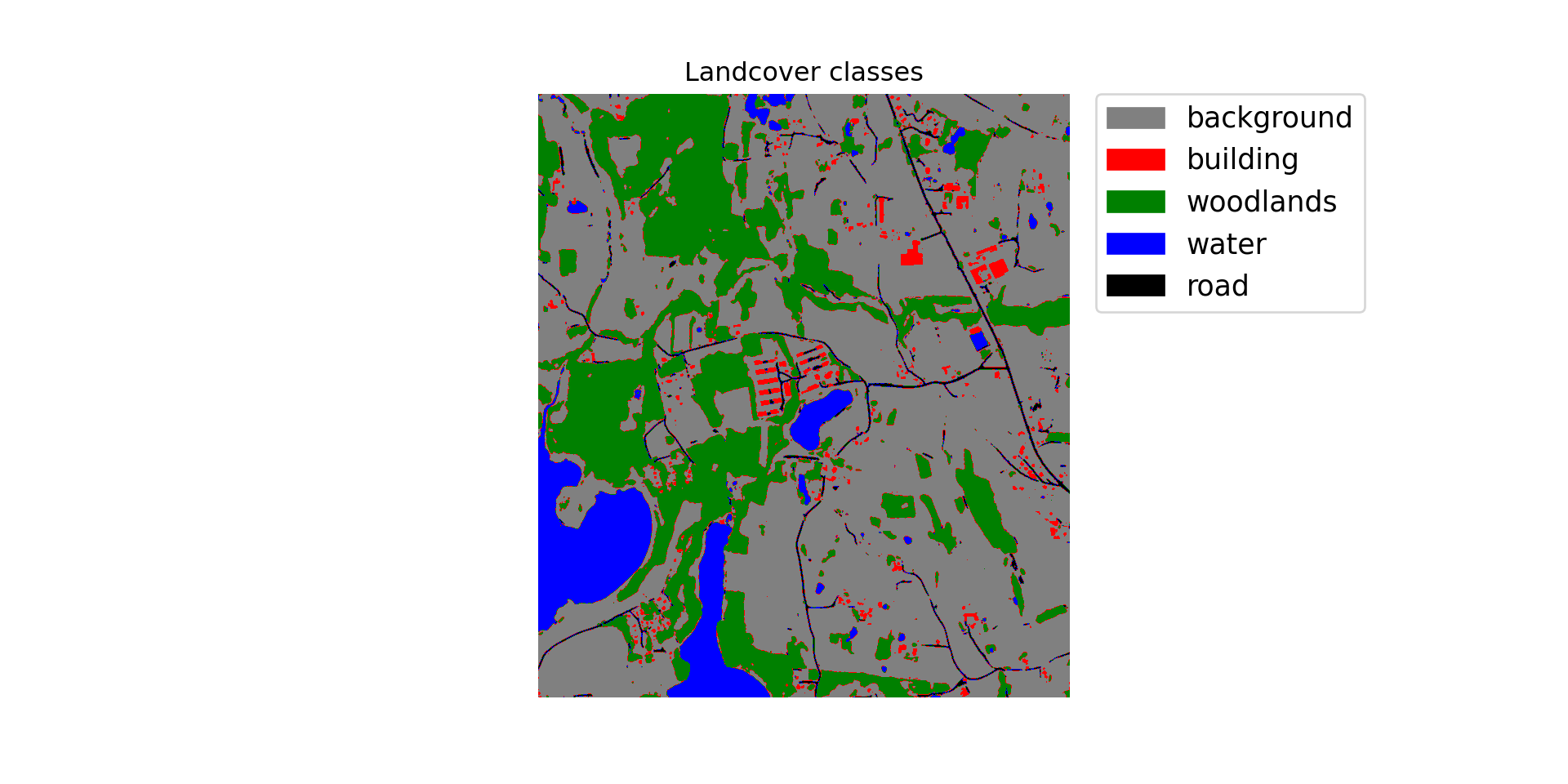
Make Probabilistic Predictions
Below I have redefined the inference function to output the predicted probability for each class as opposed to the hard classification. The result will be a multiband raster with a channel for each class. I have added a new nCls parameter which indicates the number of classes being differentiated. When predicting each chip, I do not use argmax() to obtain the index associated with the class with the highest logit. Instead, the results are passed through a softmax function to obtain class probabilities.
def geoInferProb(image_in, pred_out, nCls, chip_size, stride_x, stride_y, crop, n_channels):
#Read in topo map and convert to tensor===========================
image1 = cv2.imread(image_in)
image1 = cv2.cvtColor(image1, cv2.COLOR_BGR2RGB)
image1 = image1.astype('uint8')
image1 = torch.from_numpy(image1)
image1 = image1.permute(2, 0, 1)
image1 = image1.float()/255
t_arr = image1
#Make blank grid to write predictions two with same height and width as topo===========================
p_arr = torch.zeros((nCls, t_arr.shape[1], t_arr.shape[2])).float()
#Predict to entire topo using overlapping chips, merge back to original extent=============
size = chip_size
stride_x = stride_x
stride_y = stride_y
crop = crop
n_channels = n_channels
across_cnt = t_arr.shape[2]
down_cnt = t_arr.shape[1]
tile_size_across = size
tile_size_down = size
overlap_across = stride_x
overlap_down = stride_y
across = math.ceil(across_cnt/overlap_across)
down = math.ceil(down_cnt/overlap_down)
across_seq = list(range(0, across, 1))
down_seq = list(range(0, down, 1))
across_seq2 = [(x*overlap_across) for x in across_seq]
down_seq2 = [(x*overlap_down) for x in down_seq]
#Loop through row/column combinations to make predictions for entire image
for c in across_seq2:
for r in down_seq2:
c1 = c
r1 = r
c2 = c + size
r2 = r + size
#Default
if c2 <= across_cnt and r2 <= down_cnt:
r1b = r1
r2b = r2
c1b = c1
c2b = c2
#Last column
elif c2 > across_cnt and r2 <= down_cnt:
r1b = r1
r2b = r2
c1b = across_cnt - size
c2b = across_cnt + 1
#Last row
elif c2 <= across_cnt and r2 > down_cnt:
r1b = down_cnt - size
r2b = down_cnt + 1
c1b = c1
c2b = c2
#Last row, last column
else:
c1b = across_cnt - size
c2b = across_cnt + 1
r1b = down_cnt - size
r2b = down_cnt + 1
ten1 = t_arr[0:n_channels, r1b:r2b, c1b:c2b]
ten1 = ten1.to(device).unsqueeze(0)
model.eval()
with torch.no_grad():
ten2 = model(ten1)
m = nn.Softmax(dim=1)
pr_probs = m(ten2)
ten_p = pr_probs.squeeze()
#print("executed for " + str(r1) + ", " + str(c1))
if(r1b == 0 and c1b == 0): #Write first row, first column
p_arr[:, r1b:r2b-crop, c1b:c2b-crop] = ten_p[:, 0:size-crop, 0:size-crop]
elif(r1b == 0 and c2b == across_cnt+1): #Write first row, last column
p_arr[:,r1b:r2b-crop, c1b+crop:c2b] = ten_p[:,0:size-crop, 0+crop:size]
elif(r2b == down_cnt+1 and c1b == 0): #Write last row, first column
p_arr[:,r1b+crop:r2b, c1b:c2b-crop] = ten_p[:,crop:size+1, 0:size-crop]
elif(r2b == down_cnt+1 and c2b == across_cnt+1): #Write last row, last column
p_arr[:,r1b+crop:r2b, c1b+crop:c2b] = ten_p[:,crop:size, 0+crop:size+1]
elif((r1b == 0 and c1b != 0) or (r1b == 0 and c2b != across_cnt+1)): #Write first row
p_arr[:,r1b:r2b-crop, c1b+crop:c2b-crop] = ten_p[:,0:size-crop, 0+crop:size-crop]
elif((r2b == down_cnt+1 and c1b != 0) or (r2b == down_cnt+1 and c2b != across_cnt+1)): # Write last row
p_arr[:,r1b+crop:r2b, c1b+crop:c2b-crop] = ten_p[:,crop:size, 0+crop:size-crop]
elif((c1b == 0 and r1b !=0) or (c1b ==0 and r2b != down_cnt+1)): #Write first column
p_arr[:,r1b+crop:r2b-crop, c1b:c2b-crop] = ten_p[:,crop:size-crop, 0:size-crop]
elif (c2b == across_cnt+1 and r1b != 0) or (c2b == across_cnt+1 and r2b != down_cnt+1): # write last column
p_arr[:,r1b+crop:r2b-crop, c1b+crop:c2b] = ten_p[:,crop:size-crop, 0+crop:size]
else: #Write middle chips
p_arr[:,r1b+crop:r2b-crop, c1b+crop:c2b-crop] = ten_p[:,crop:size-crop, crop:size-crop]
#Read in a GeoTIFF to get CRS info=======================================
image3 = rio.open(image_in)
profile1 = image3.profile.copy()
image3.close()
profile1["driver"] = "GTiff"
profile1["dtype"] = "float32"
profile1["count"] = nCls
profile1["PHOTOMETRIC"] = "MINISBLACK"
profile1["COMPRESS"] = "NONE"
pr_out = p_arr.cpu().numpy().astype('float32')
#Write out result========================================================
with rio.open(pred_out, "w", **profile1) as f:
f.write(pr_out)
torch.cuda.empty_cache()I now execute the new function for the input image. You can open each band in the resulting raster grid using your GIS software to explore the predicted class probabilities.
rpbOut = geoInferProb(image_in=testImg,
pred_out="data/files/example_pred_prob.tif", nCls=5,
chip_size=512, stride_x=256, stride_y=256, crop=50, n_channels=3)Concluding Remarks
You can now use trained models to make predictions back to geospatial data using a custom Python function that allows larger extents to be processed as overlapping image chips and subsequently reassembled to create a contiguous output. In the next section, we will explore a set of R functions that my lab group created to generate raster masks, image chips, lists of image chips, and associated statistics.